Rows: 14,845
Columns: 19
$ id <chr> "PSP19.3_2014-04-01_mytilus", "PSP21.09_2014-04-01_myti…
$ location_id <chr> "PSP19.3", "PSP21.09", "PSP21.2", "PSP25.17", "PSP26.07…
$ date <date> 2014-04-01, 2014-04-01, 2014-04-01, 2014-04-02, 2014-0…
$ species <chr> "mytilus", "mytilus", "mytilus", "mytilus", "mytilus", …
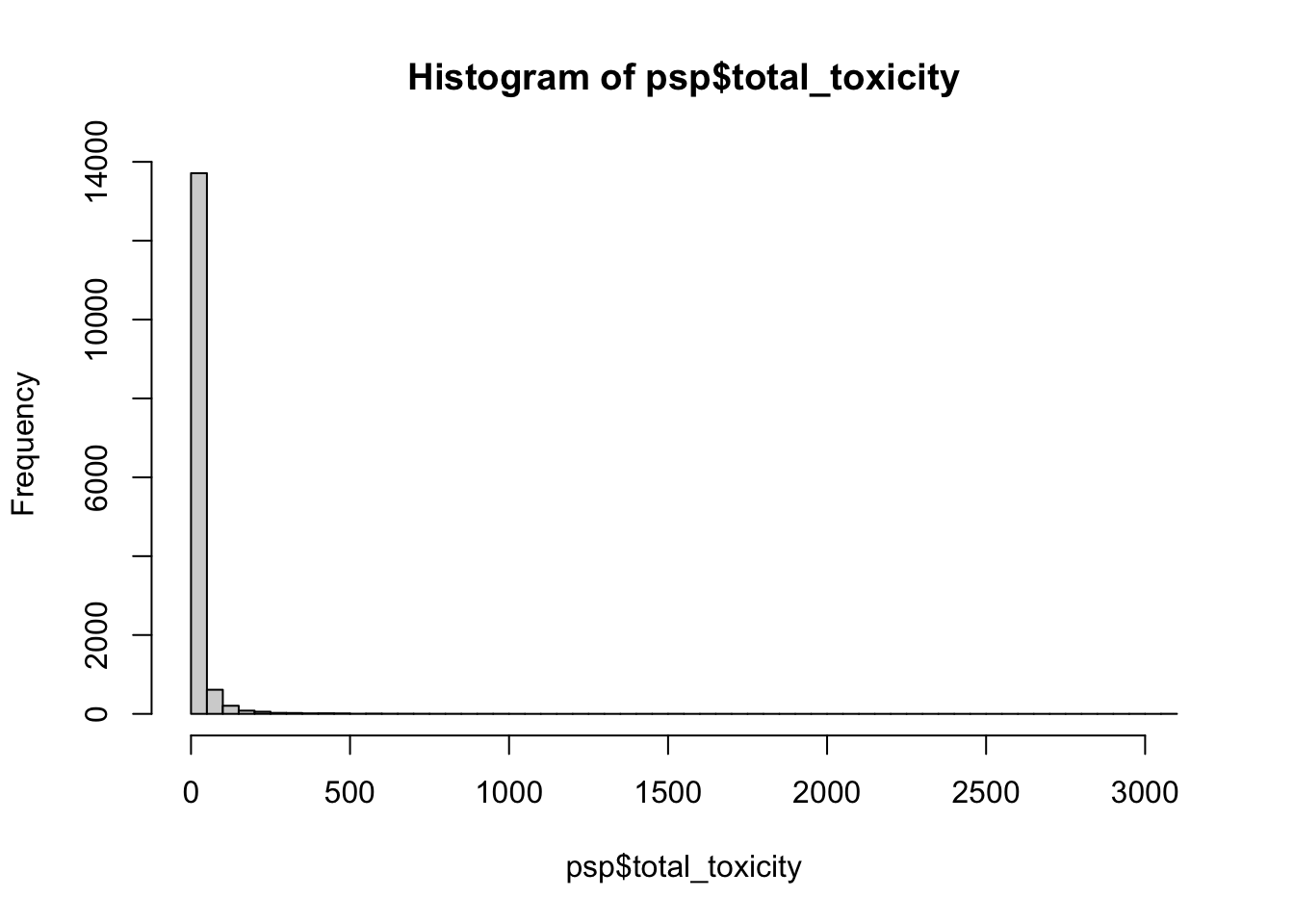
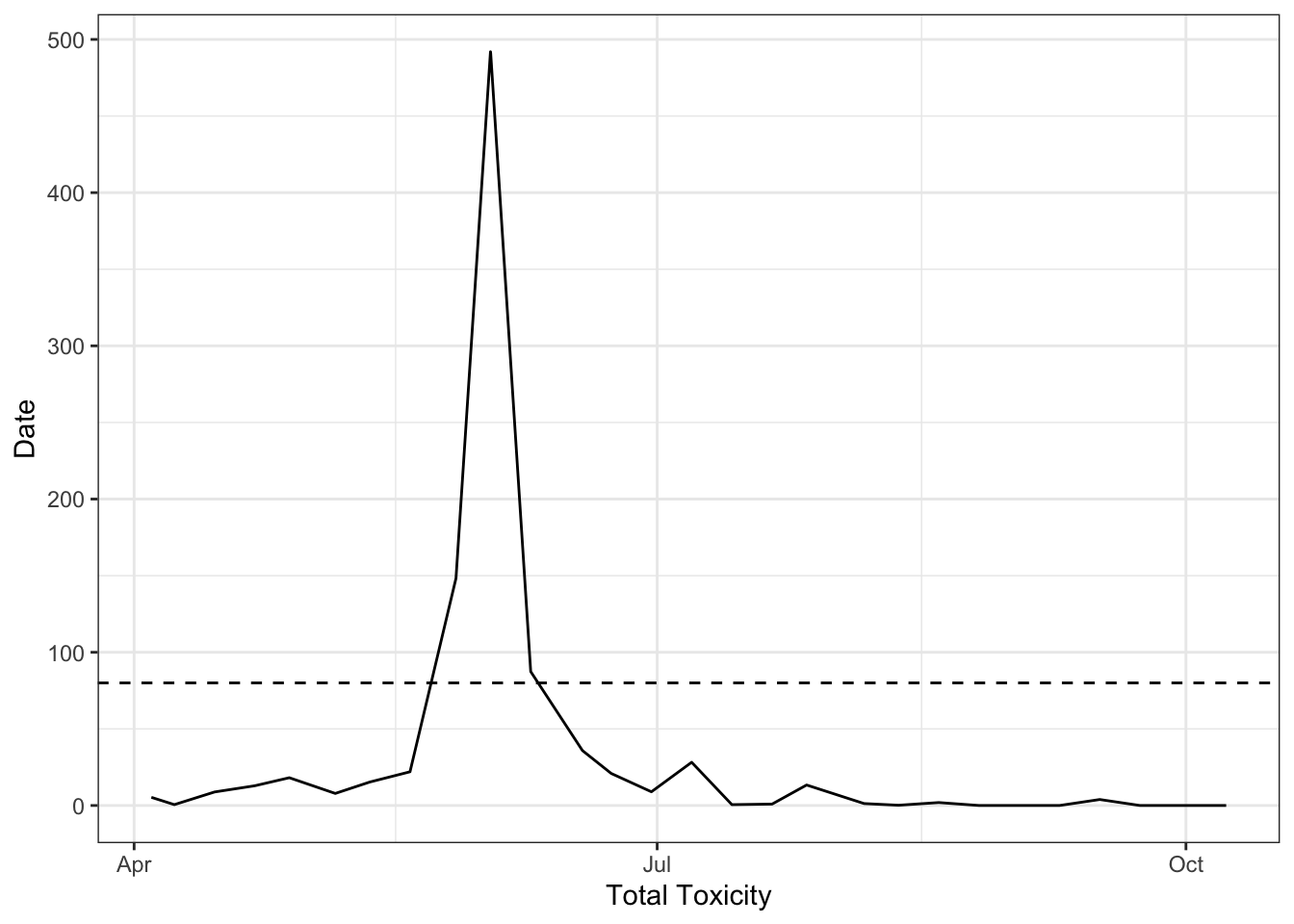
$ total_toxicity <dbl> 0.1561590, 0.2044939, 0.1933397, 0.1301325, 0.1859036, …
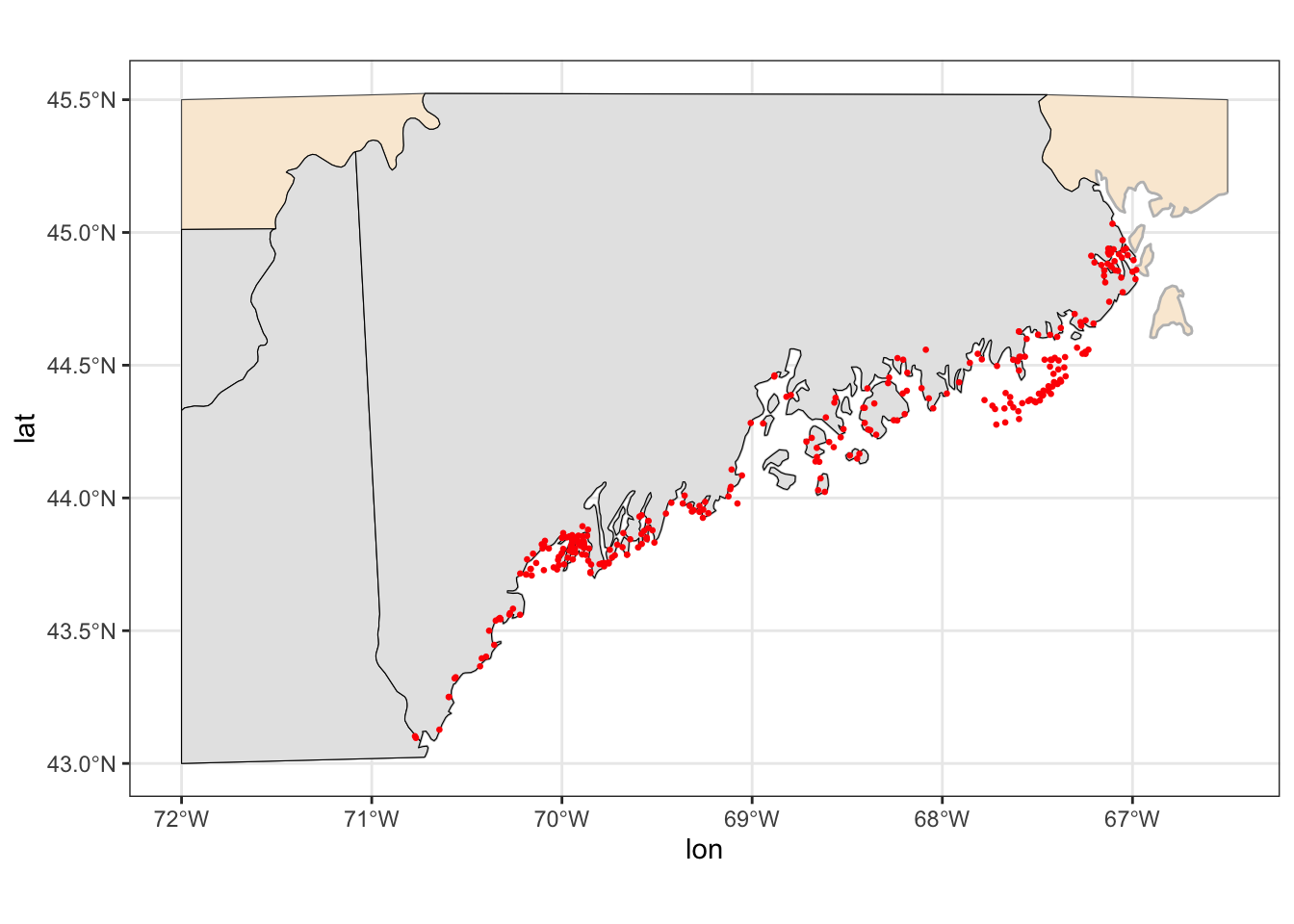
$ lat <dbl> 44.22853, 44.23824, 44.29200, 44.61438, 44.65701, 44.53…
$ lon <dbl> -68.53441, -68.34792, -68.23696, -67.43323, -67.20525, …
$ gtx4 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ gtx1 <dbl> 0.000000, 0.000000, 0.000000, 0.000000, 0.000000, 0.000…
$ dcgtx3 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ gtx5 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ dcgtx2 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ gtx3 <dbl> 0.1561590, 0.2044939, 0.1933397, 0.1301325, 0.1859036, …
$ gtx2 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ neo <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ dcstx <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ stx <dbl> 0.000000, 0.000000, 0.000000, 0.000000, 0.000000, 0.000…
$ c1 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…
$ c2 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0…